Research: Machine Learning in Cancer Genomics
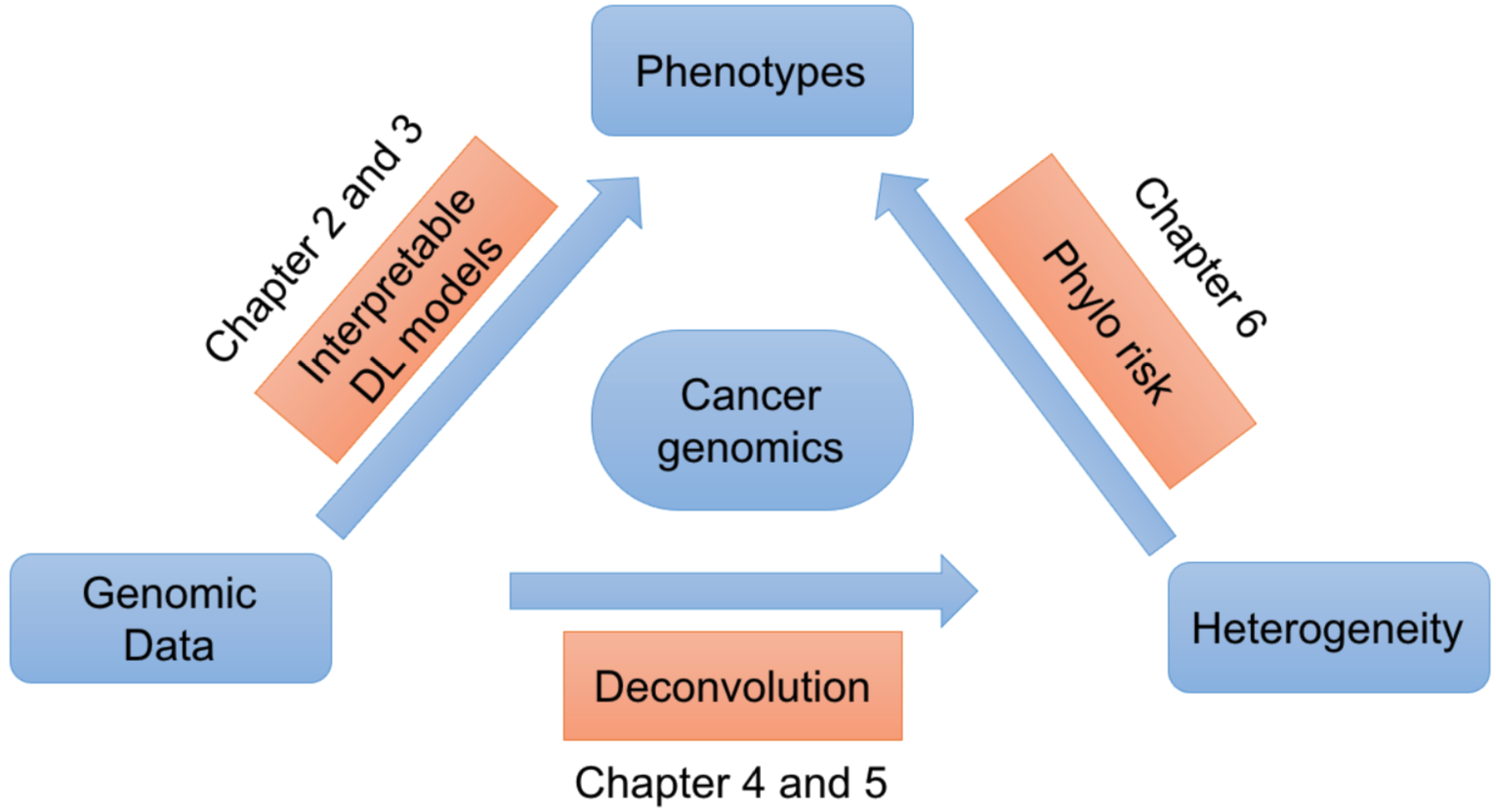
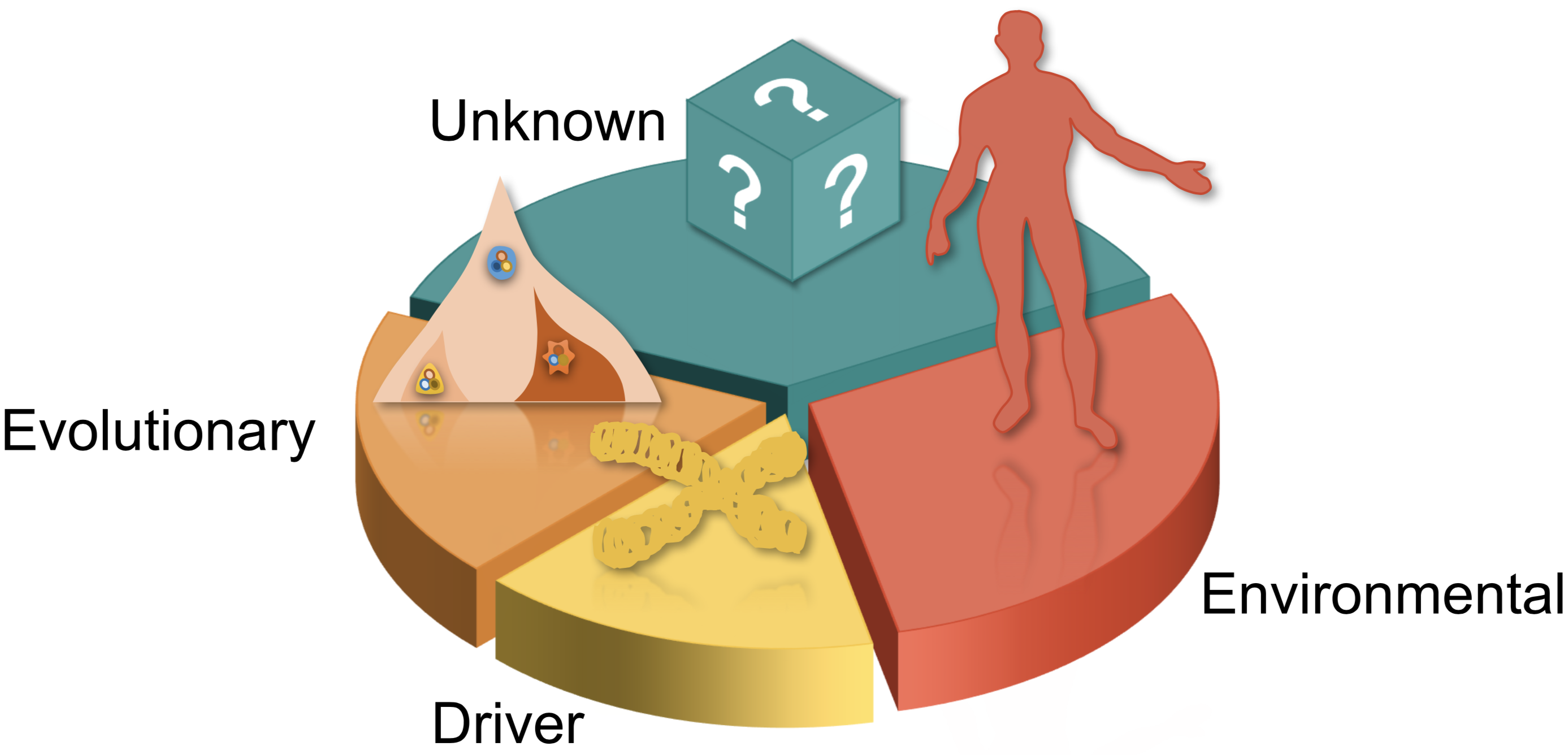
Cancer proceeds from the accumulation of genomic alterations, and develops into heterogeneous cell populations in an evolutionary process. Therefore, the prognoses of cancer patients, such as survival profile, metastasis, and drug response, are encoded by the large-volume genome data. Our research focuses on the personalized medicine of cancer with machine learning and phylogenetic models (
Thesis;
Book):
- Reliable phenotype inference of cancer through well-designed interpretable machine learning models.
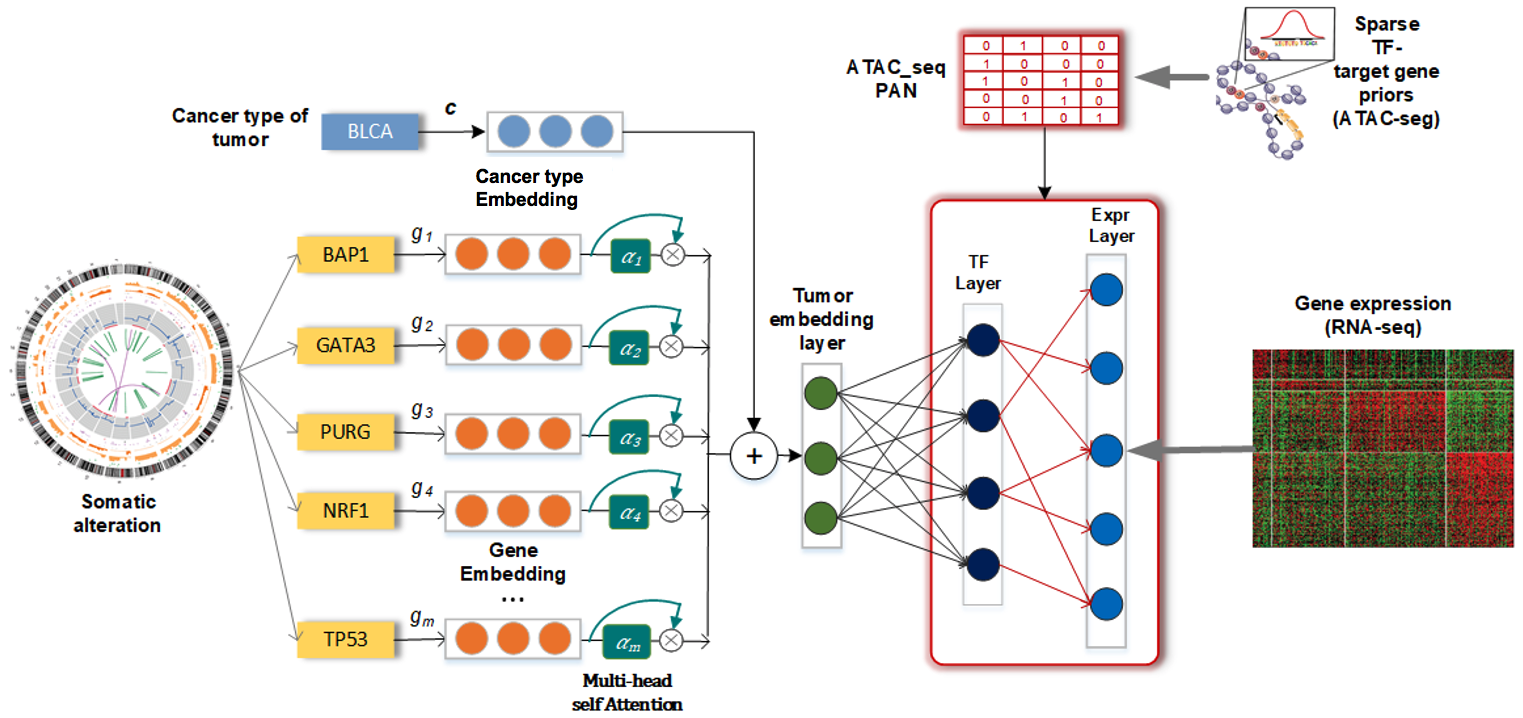
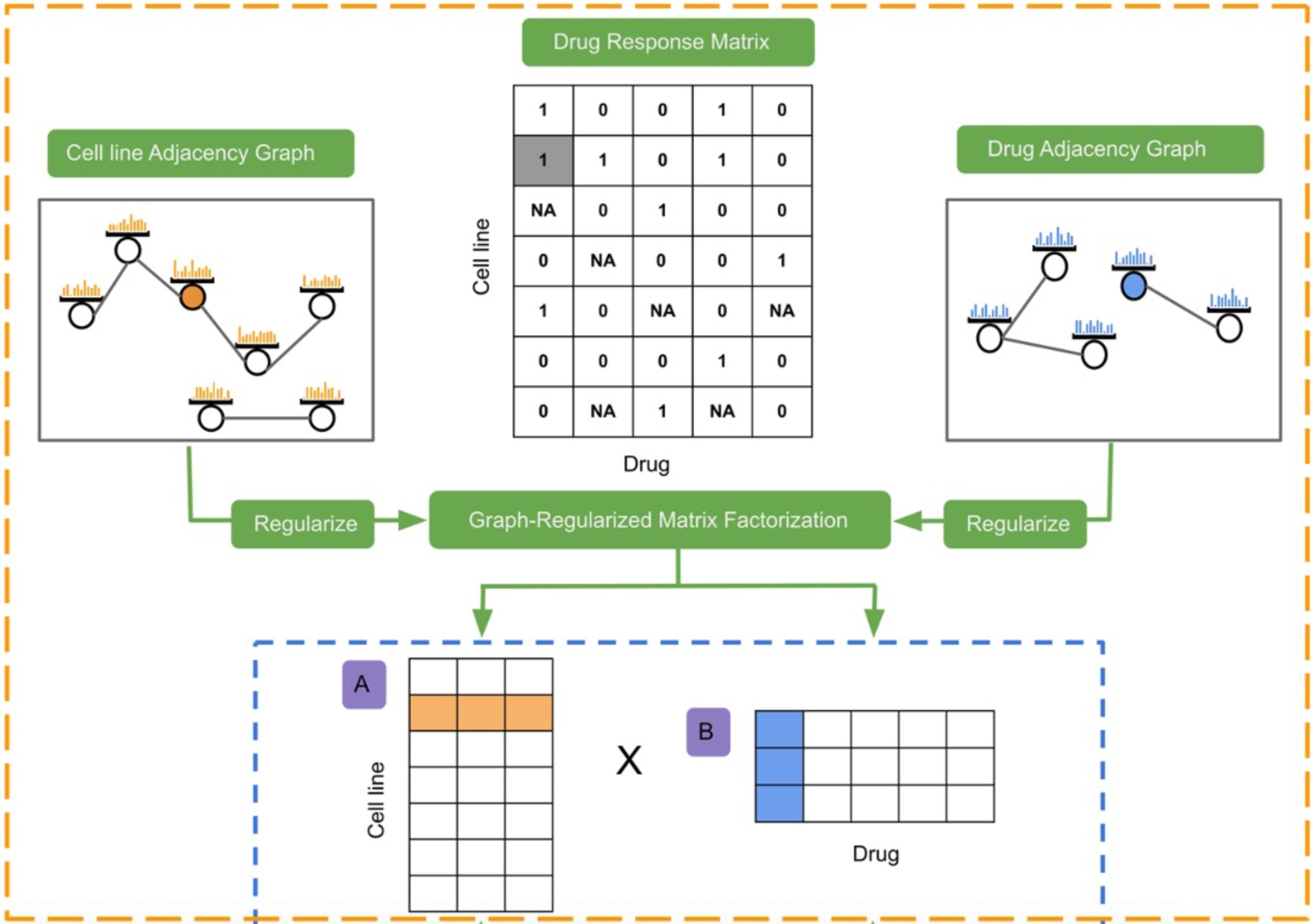
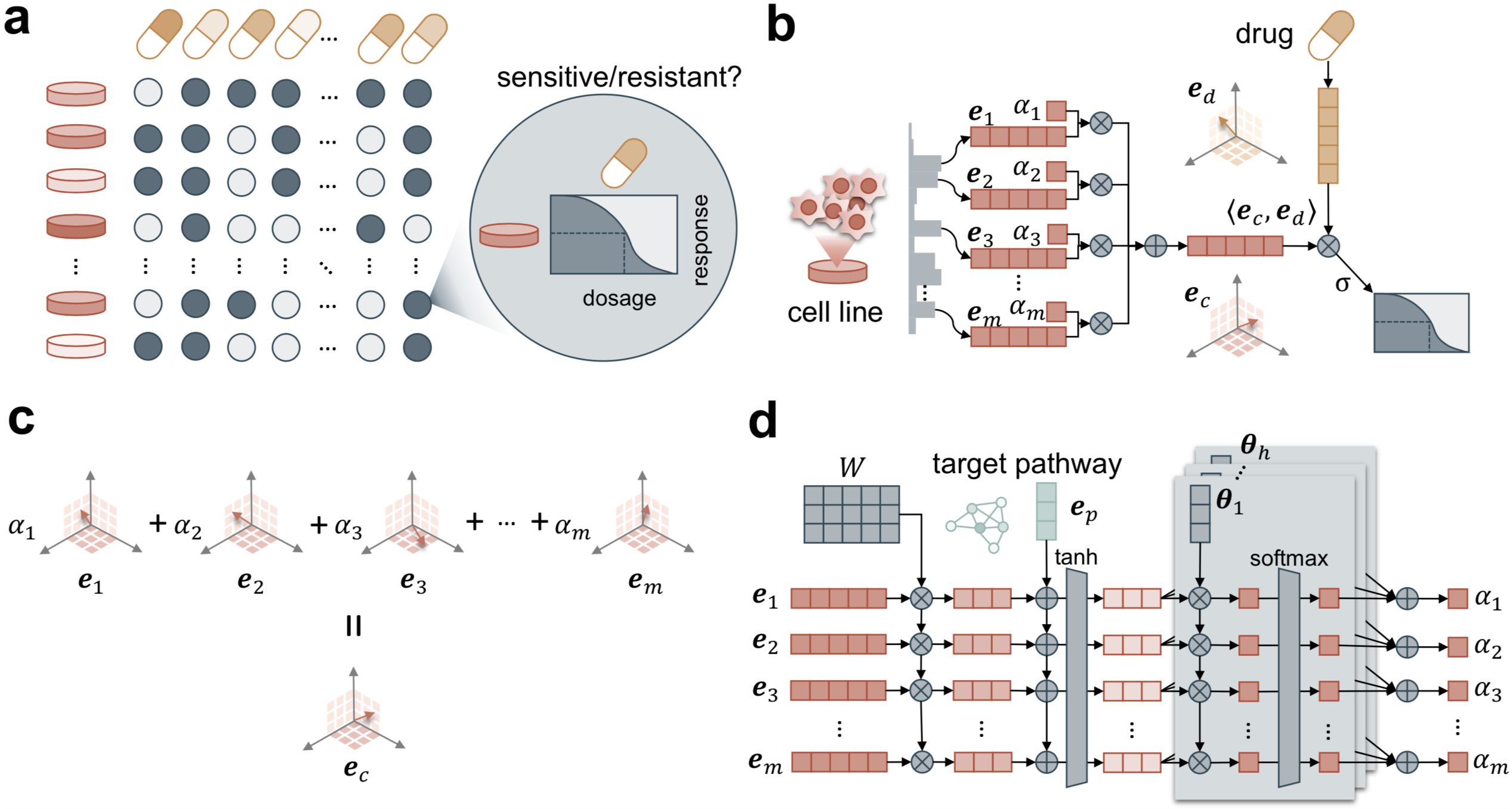
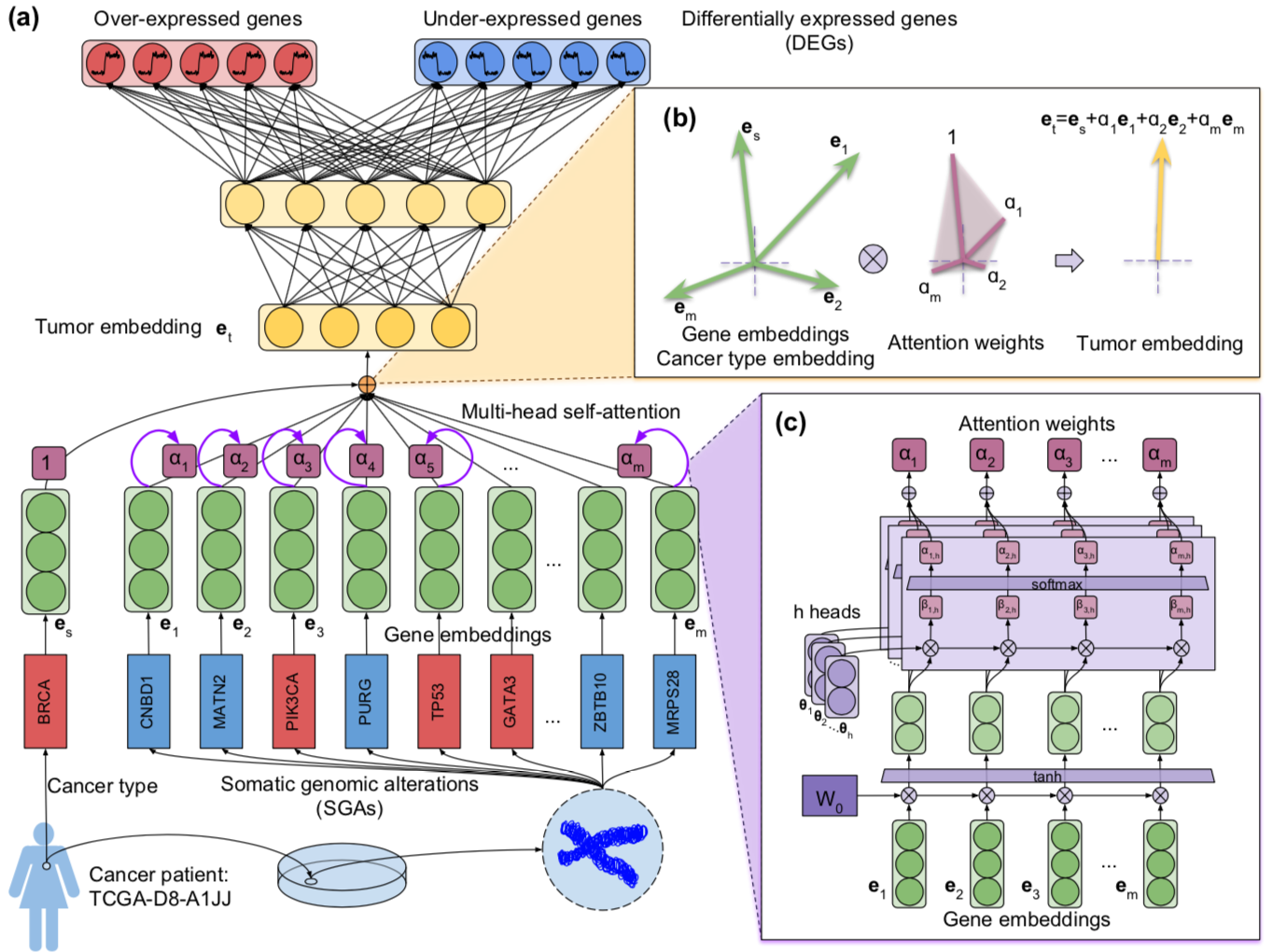
By leveraging the power of large scale genomic data and external biomedical knowledge base, we have been working on deep learning models for the accurate inference of cancer phenotypes, including transcriptome expression levels (Genomic Impact Transformer: GIT), transcription factor activities (Chromatin-informed Inference of Transcriptional Regulators Using Self-attention mechanism: CITRUS), and drug resistance (Contextual Attention-based Drug REsponse: CADRE; Deep Learning-based Graph Regularized Matrix Factorization: DeepGRMF). We addressed the interpretability of models through techniques such as attention mechanism to identify driver mutations and critical biomarkers.
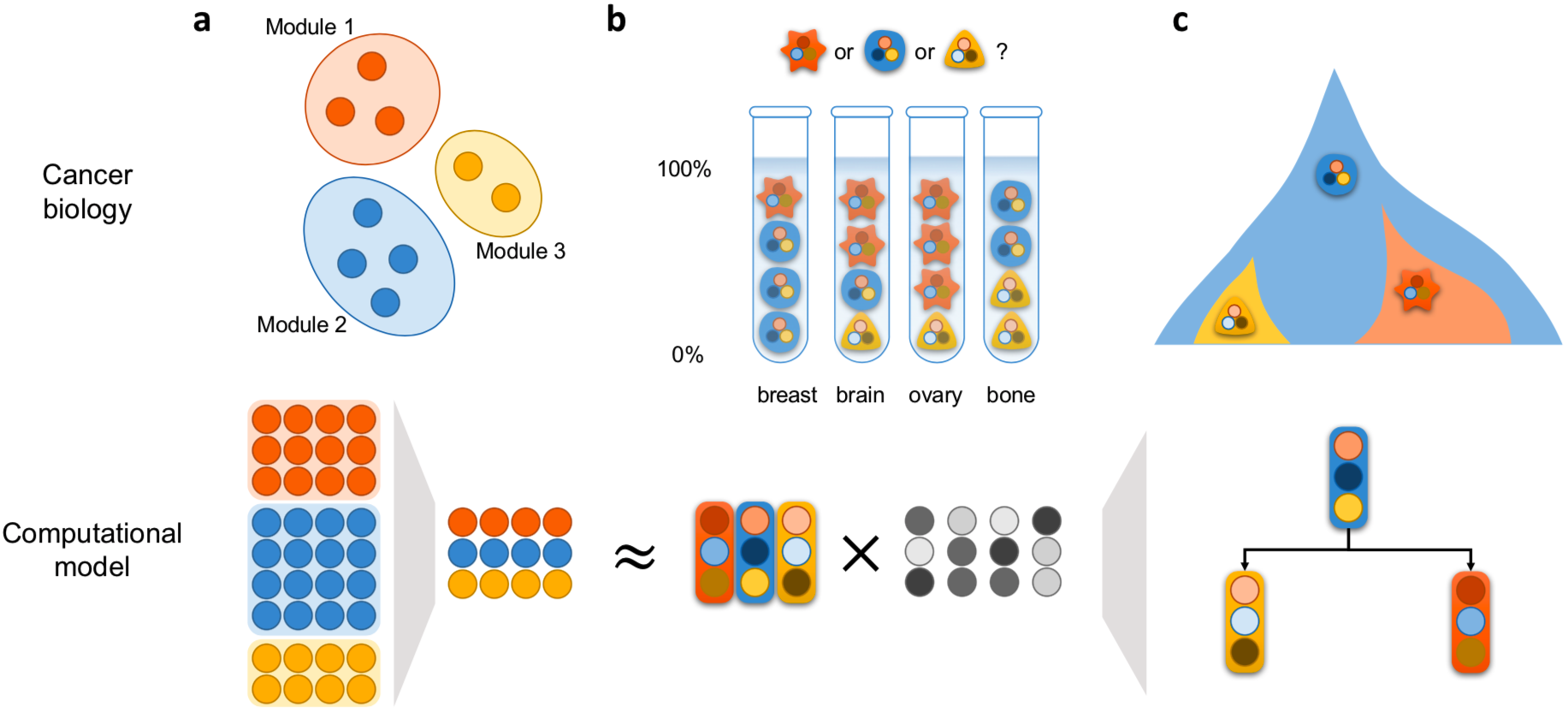
- Revealing intra-/inter-tumor heterogeneity and mechanism of tumor progression via robust deconvolution and phylogenetic algorithms.
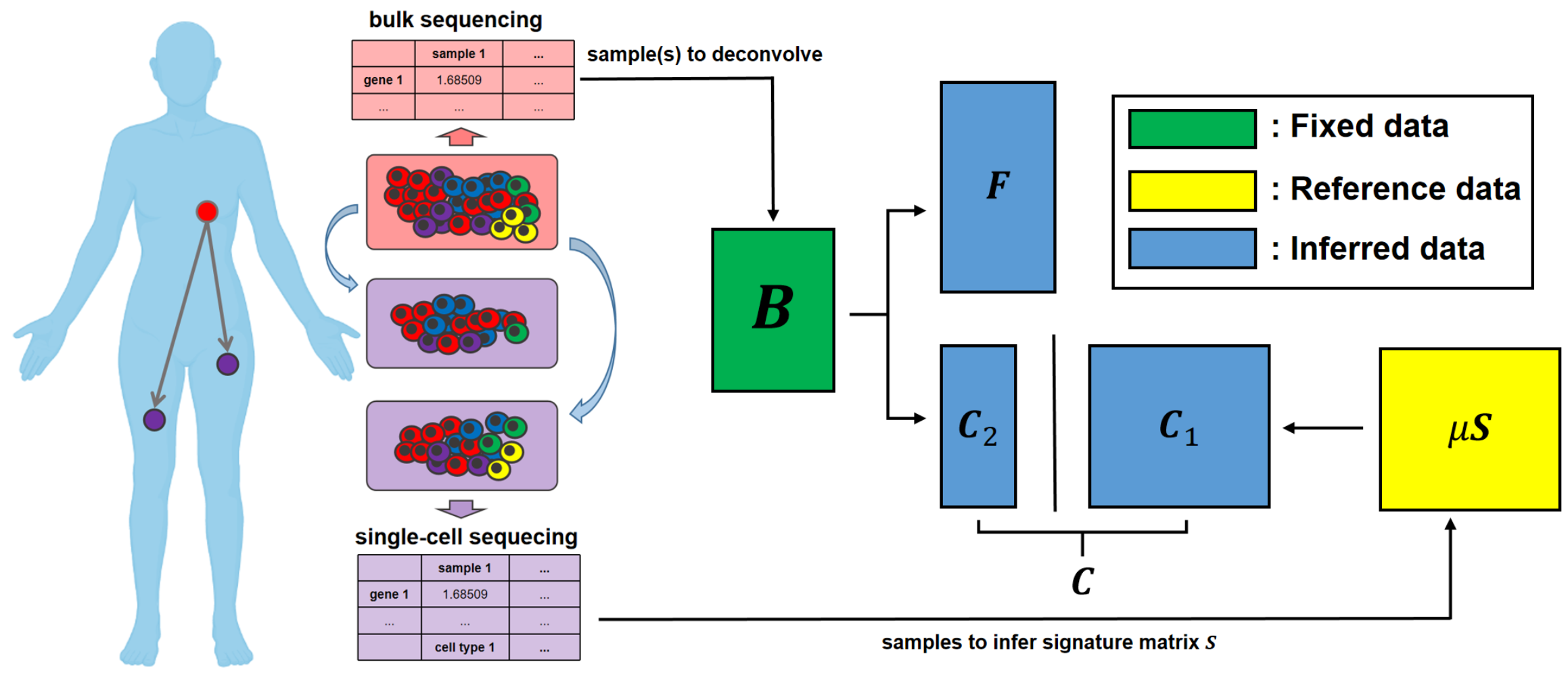
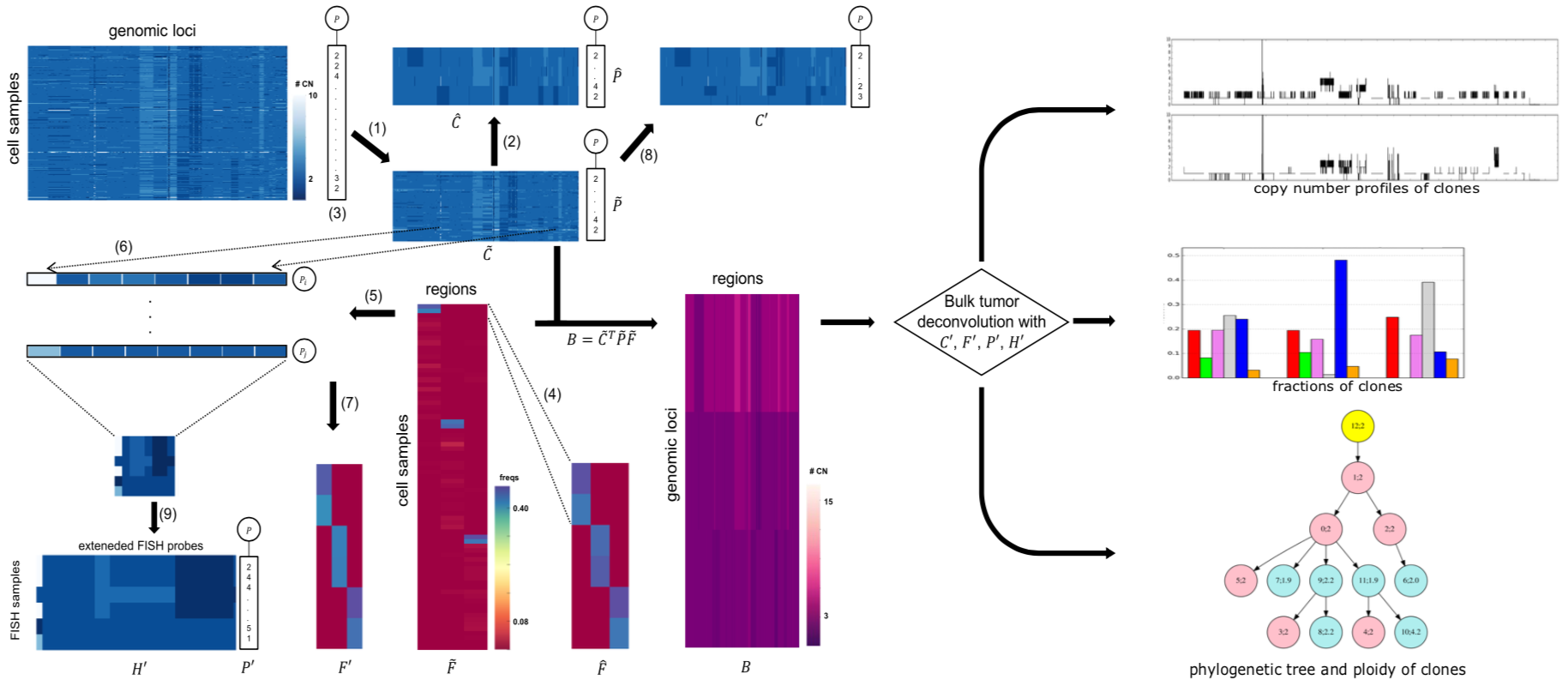
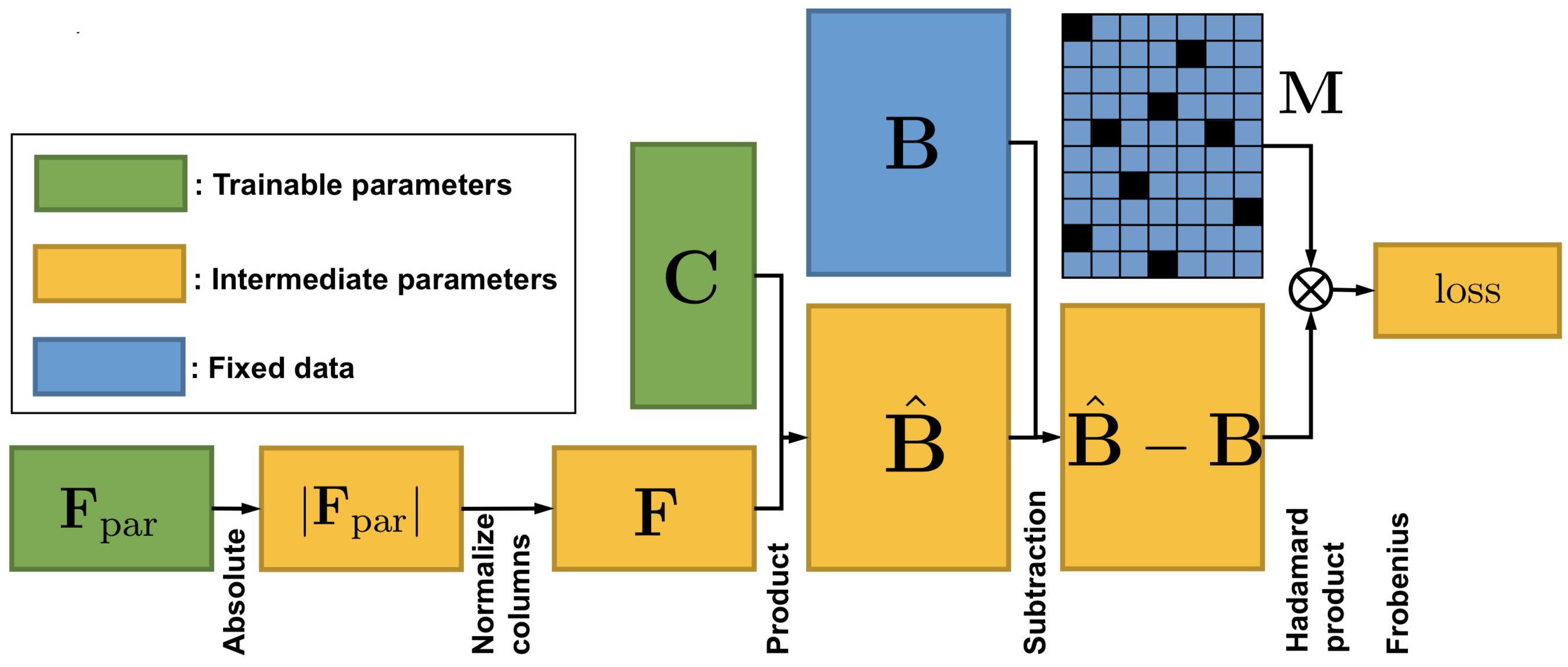
We formulated the deconvolution of bulk tumor molecular data mathematically as a biologically inspired matrix factorization problem, and proposed a neural network (Neural Network Deconvolution: NND) and then an improved hybrid optimizer (Robust and Accurate Deconvolution: RAD; RAD with single-cells: RADs) to solve the problem robustly and accurately.
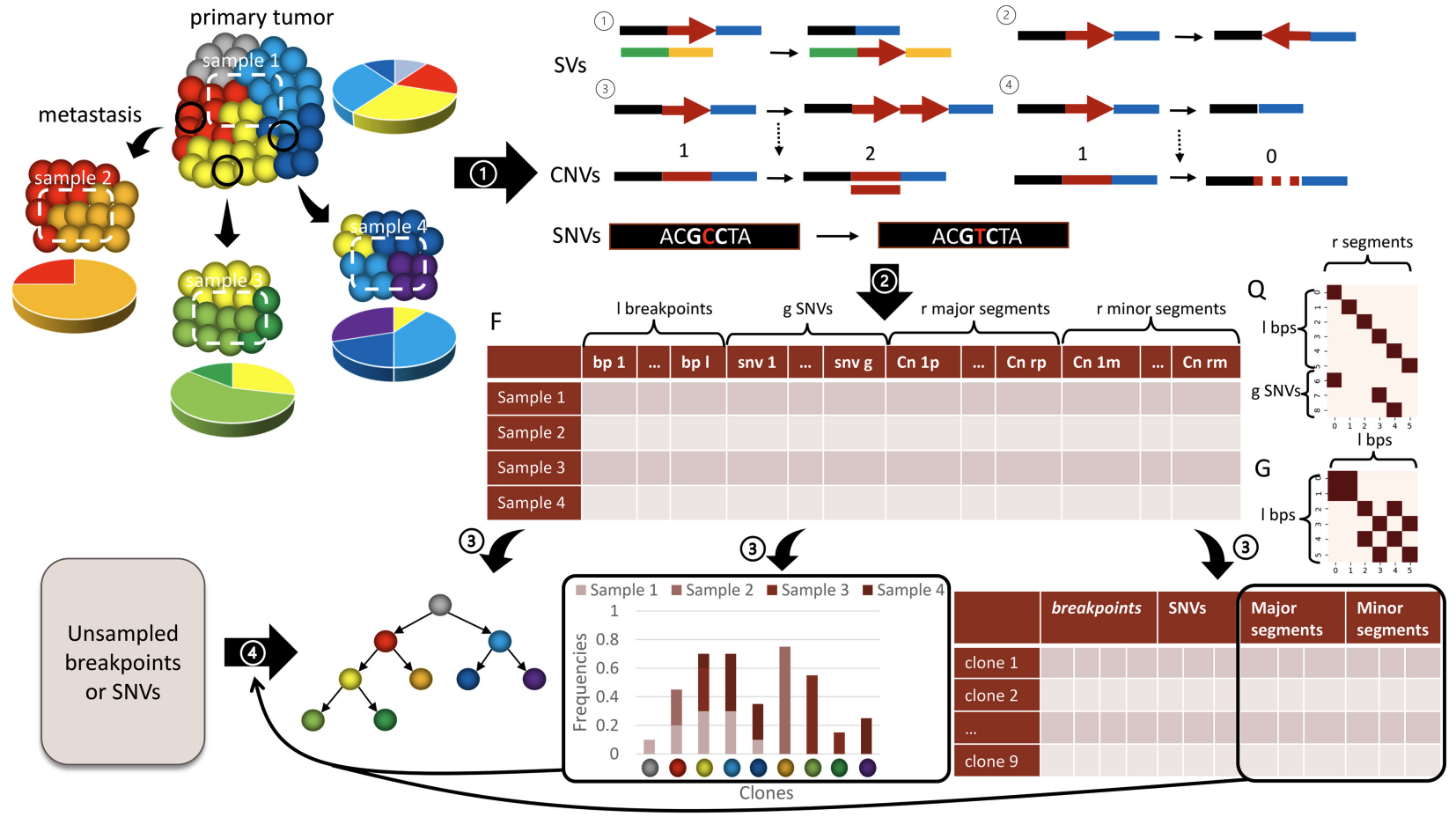
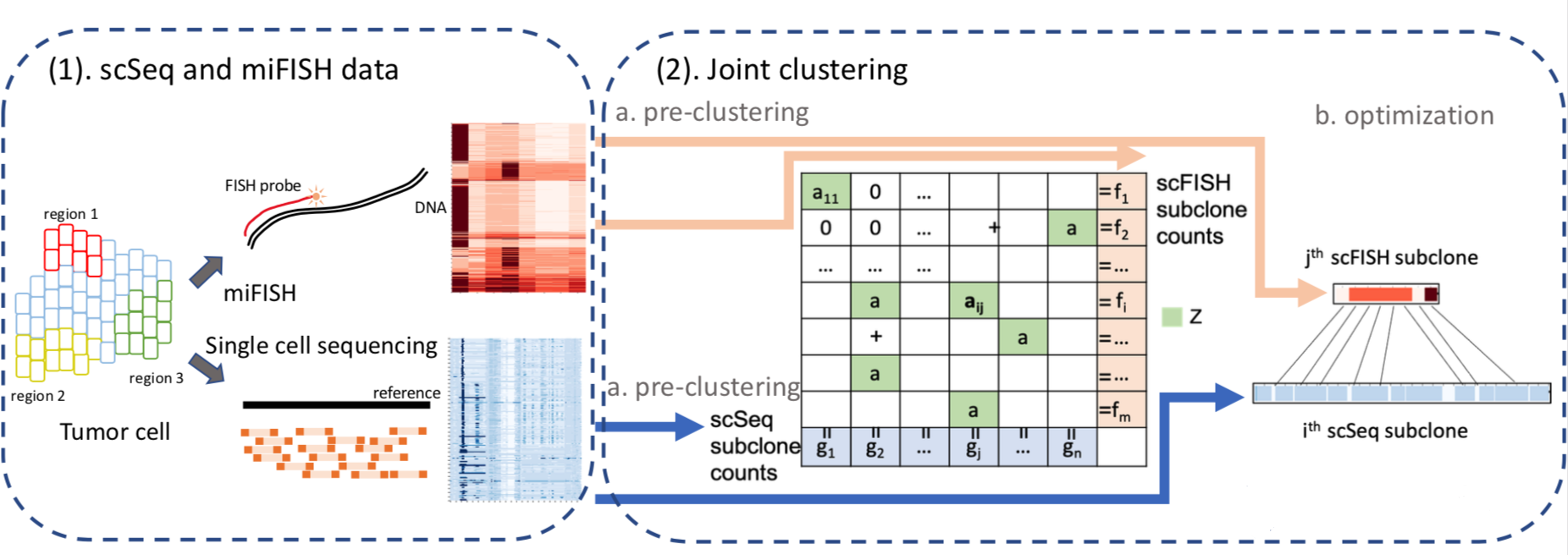
We developed and applied a Minimum Elastic Potential (MEP) algorithm to reconstruct the evolutionary trajectory from the unmixed clones. We utilized mixed integer linear programming to solve deconvolution from bulk genomic data (FISH-Deconv; Joint-Clustering; TUSV-ext).
- Improving prognostic prediction of cancer by incorporating machine learning and evolutionary methods.
Clinicians traditionally focused on the pathological features and driver-level genomic profiles to facilitate the treatment. However, it is possible that critical clones, instead of the bulk tumor as a whole, affect the prognoses. We explored the questions by integrating both the evolutionary mutational features, driver-level features, and clinical features to improve the prognostic prediction of cancer. We developed an L0-regularized Cox regression model (Phylo-Risk), and found that the evolutionary features account for roughly 1/3 of all the available features, depending on cancer types and sequencing techniques.